|
You are here: MIT OpenLabWare » Aflatoxin
» Module Overview
Module Overview
Aflatoxins are a group of related compounds produced by the fungus Aspergillus flavus, a soil organism that
can grow on peanuts, corn, wheat, rice and many other basic food products. One of the compounds produced by this mold is
aflatoxin B1 (AFB1), a potent liver toxin and carcinogen. For more information on how
AFB1 was discovered and how it came to be the focus of many research groups around the world, see the video
Aflatoxin's
Story.
One of the first responses to any new toxin is to determine its chemical structure, which often yields information necessary to elucidate the compound’s mechanism of action. This was done for AFB1 as well as its cousins.
Most chemicals that cause cancer interact with DNA, and it was reasonable to speculate in the 1960’s and 1970’s that some type of DNA interaction might initiate liver cells along the pathway toward malignancy. For example, the bound forms of chemicals on DNA could result in a change in the coding properties of DNA, and the resultant mutations might cause the cell to take on the malignant phenotype. The Wogan and Büchi groups knew little about the aflatoxin molecule’s interaction with DNA. There were two general possibilities: intercalation and a chemical covalent bond forming reaction, and they were not mutually exclusive. In addition, if both were occurring, the next question was: which interaction (if either) causes liver cancer?
Comparing toxicity data of aflatoxin B1 with a related fungal metabolite, aflatoxin B2, was very helpful in determining what interactions were occurring between aflatoxin and DNA.
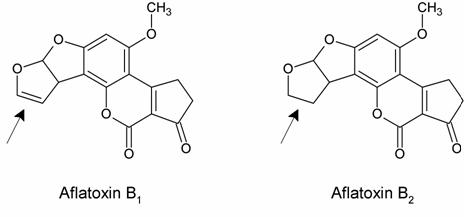
Wogan had shown that aflatoxin B1 is carcinogenic but that aflatoxin B2 is barely so. The only difference between the molecules is indicated with an arrow. Because this change does not change the molecule’s planar ring structure, it is likely that both aflatoxins intercalate with DNA. Therefore aflatoxin B1’s carcinogenicity is probably due to its covalent interactions with DNA. It was also possible that aflatoxin B1 did not react directly with DNA, but rather was transformed into a related compound through metabolism. This “activated metabolite” might instead be the carcinogenic substance that reacted with DNA.
The following reaction scheme was the working hypothesis when John Essigmann started working on the project with
Wogan and Büchi in 1976:
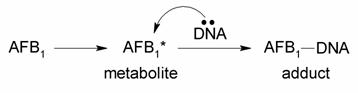
The researchers hypothesized that some nucelophilic site on the DNA molecules were reacting with AFB1*. There were two hypotheses at the time as to the nature of AFB1*, the proposed chemically reactive intermediate of aflatoxin. The first, proposed by H.L. Gurtoo, was that AFB1* was in fact aflatoxin B2a, a known abundant metabolite of AFB1. Its structure is shown below:
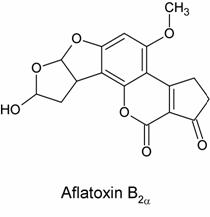
In this theory, the carcinogenic effects of AFB2a were suspected to arise from its
hemiacetal functional group, which theoretically enables it to undergo the following reaction in alkaline
solution:
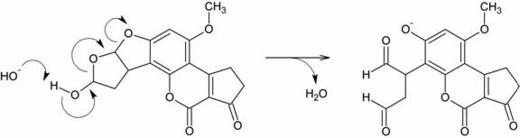
The product is a dialdehyde phenolate anion. It contains two aldehyde groups, which are quite reactive toward good nucleophiles. Purines (for example, guanine) are fairly good nucleophiles and thus it was suspected that they could react with one of the aldehydes to form a Schiff base (or, in the IUPAC nomenclature, imine) adduct as shown below:
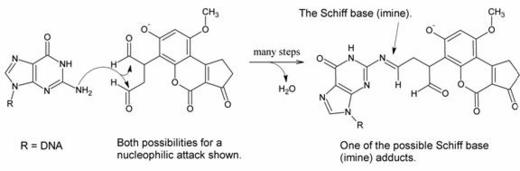
The researchers did not expect that both aldehydes would react; in their model it was either one or the other.
The second hypothesis, proposed by James and Elizabeth Miller, was that AFB1* was an epoxide. Its structure is shown below:
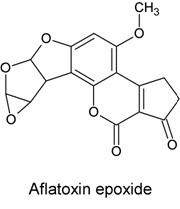
In this hypothesis, a nucelophilic site on DNA would attack one of the epoxide carbons to form the adduct, as in the reaction scheme below:
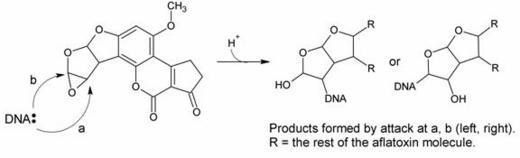
There was evidence both for and against this hypothesis. Evidence in favor included the fact that, upon acid hydrolysis of the adduct, a species known as the dihydrodiol (shown below) was formed:
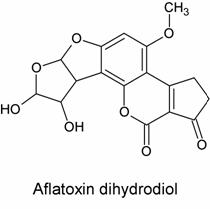
This product is consistent with hydrolysis of the proposed epoxide-formed adducts above but is not consistent with hydrolysis of the Schiff base (imine) adduct mentioned previously. Evidence against this hypothesis was much less solid but nevertheless significant: no one had been able to synthesize the epoxide, despite many attempts, or even prove its existence.
John Essigmann’s thesis project as a graduate student in Wogan’s lab was to define chemical modifications of nucleic acids that occurred during the process of carcinogenesis. The aflatoxin adduct identification project began as the last part of his doctoral work. For more information about John’s background, watch the following series of videos (they are in chronological order):
The last video describes two important chemical pieces of information concerning aflatoxin that turn out to be significant later on.
A number of different obstacles and problems had to be overcome to determine the adduct structure, and the experiments in the timeline section can be sorted by these subsections, known as “paths.” They are:
- Modification of DNA using aflatoxin
The first step was to make the adduct (if in fact the carcinogenic process stemmed from an adduct; for additional
hypotheses, see work by W.
Thilly in Nature Genetics), and a method to do this in vitro had previously been worked out in the laboratory
by W. Busby. So, most of the experiments in this section are incubations of aflatoxin with DNA and the development of an
enzymatic system that formed AFB1*.
- Liberation of adduct from modified DNA
Once the putative adduct had been formed, the next task was to remove it from DNA using hydrolysis methods. A
good introduction to this topic is the video Hydrolysis
Methods.
(Be sure to watch The
Early Meeting with George
Büchi first.)
- Isolation of liberated adduct using the extraction approach
The next theoretical step is to isolate the adduct from the hydrolysate (the hydrolyzed modified DNA). This short set
of experiments attempted using a simple extraction to isolate the adduct.
- Isolation of liberated adduct using the precolumn approach
This alternative to extraction attempted to isolate the adduct using a precolumn. (Good chromatography resources can be found at http://www.chromatography-online.org/index.html. A working knowledge of the
principles underlying chromatographic systems is essential to understand this and the following path.)
- Isolation of large (mg) quantities of adduct using the preparative column approach
Isolating enough adduct to do structural determination was an enormous obstacle. At the time, quantities on the order of milligrams were needed to characterize a structure. A mistake (and the ability to turn it to one’s advantage) led to the isolation of a large quantity of pure adduct.
- Preliminary structure determination
Several spectroscopic techniques were used to determine the preliminary structure of the adduct. As with the previous path, a good
understanding of spectroscopy is necessary to appreciate fully the work done in deducing the structure. An excellent spectroscopy tutorial can be
found at http://www.cem.msu.edu/~reusch/VirtualText/Spectrpy/spectro.htm.
- Determination of guanine attachment site to adduct
Once the preliminary structure had been formulated, one question remained: on which atom was the guanine attached to the aflatoxin? A beautiful series of experiments yielded a solid answer.
- Miscellaneous or unknown
- Publication or presentation
The data were in; all that was left was to publish. Was the group quick enough to be the first group with the adduct structure?
There are some facts that are essential to keep in mind when reading certain sections of the timeline or viewing some of the videos. These are:
- The radioactivity is due to tritium (3H). The aflatoxin molecule is radiolabeled in its cyclopentenone ring.
- Aflatoxin absorbs light of wavelengths 365 and 254 nm, while the DNA bases absorb only at 254 nm.
- A “precolumn” is not the same as a “preparative” or “prep” column. The former is a short column placed ahead of the analytical column to ‘protect’ it; the latter is a much longer, wider column used exclusively to isolate large quantities of material.
|